Uracil
Contents
Explanation
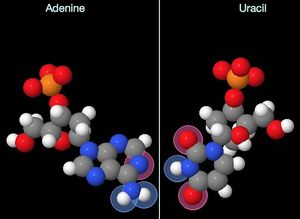
Uracil is found in RNA, the DNA-like molecule involved in copying genetic information for translation (mRNA) as well as making up several of the participants in translation (tRNA, rRNA). Its pairing partner is adenine, with which it forms two hydrogen bonds.
Frequently Asked Questions
Why did evolution 'switch' to the use of thymine for every DNA-using organism we know of?
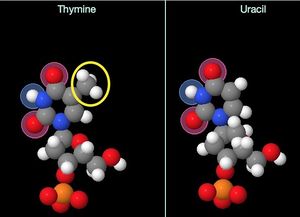
The answer is a bit complicated; it actually starts with cytosine. Cytosine can undergo a very common chemical reaction with water that results in changes to two of its basepairing positions. However the shortest summary of what happens is that this reaction changes cytosine to uracil. Literally. There is no chemical difference between a 'real' uracil and a cytosine that has had a chance interaction with water and become uracil.
How does DNA using thymine instead of uracil help?
Treating the bases as letters helps: in RNA, the code letters are A, G, C, U. If a C => U change occurs, then nothing 'looks' wrong to the machines inspecting the RNA molecule for errors—uracils are normal parts of the code. But in DNA, code letters are A, G, C, T. If a C becomes a U, it's instantly obvious—there are no (legitimate) "U's" in DNA. On this basis, mutations arising from C => U mutations can be 'spotted' and prevented in DNA... but in RNA, there's nothing that can be done.
